Mechanisms of RNA binding by LARP6
The major project in the lab is to characterize the molecular mechanism by which the RNA-binding protein LARP6 binds to its RNA targets. This work uses multiple approaches to explore the functional evolution of the LARP6 family of post-transcriptional regulators. We hypothesize that individual domains and motifs within LARP6 interact with one another to coordinate the specific RNA binding that dictates the physiological function of LARP6.
We are using comparative phylogenetics to evaluate how the LARP6 protein and its endogenous ligands have co-evolved over time. To do this, we have recombinantly expressed and purified the LARP6 proteins from several species of fish, including Xiphophorus maculatus (platyfish) and Danio rerio (zebrafish). These proteins have significant sequence similarity to the human LARP6 protein, but there are several regions that show considerable sequence divergence. We are testing whether these evolutionary sequence changes confer differences in structural stability or RNA binding activity.
Our initial studies were recently published in Protein Expression and Purification, and describe a novel intramolecular interactions between the N-terminal domain and the La module. We are now studying both the N-terminal and C-terminal domains of several LARP6 proteins, to evaluate their contribution to LARP6 structure and function.
We are also interested in identifying the molecular determinants of specific binding by LARP6. These determinants include the sequence and structures that define a LARP6 ligand, as well as the sequence and structural elements of LARP6 that create functional binding site or sites for RNAs. In particular, we are studying how multiple domains of LARP6 work together to coordinate RNA binding and cellular localization by evaluating the strength of nuclear localization and export sequence motifs and their contribution to RNA binding activity.
This project is supported by the National Institutes of Health – National Institute of General Medical Sciences through an AREA grant (GM119096).
Role of Phase Separation in Nucleic Acid-Binding Protein Function
collaboration with Steven Whitten (TxSt Department of Chemistry and Biochemistry), Loren Hough (University of Colorado Boulder), Nick Fitzkee (Mississippi State University), and Jack Correia (University of Mississippi Medical Center)
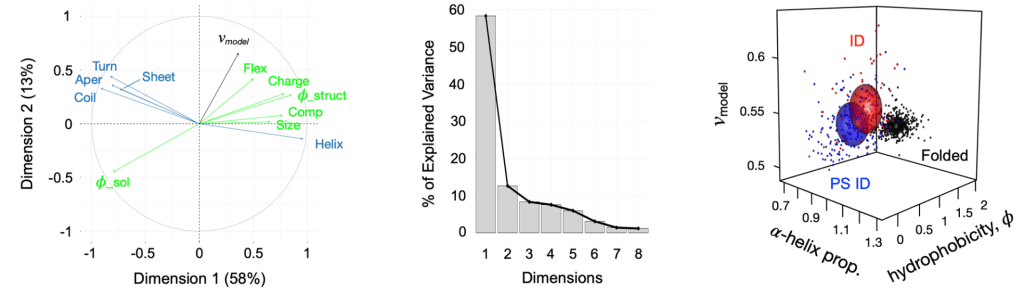
A multi-institutional collaboration led by the Whitten Lab recently developed ParSe, a computational algorithm that accurately predicts intrinsically-disordered regions of proteins that are likely to undergo liquid-liquid phase separation. A ParSe analysis of the human proteome yielded several predictions of LLPS in proteins that were not previously identified as phase-separating. We are testing some of these predictions both in vitro and in cellulo, focusing on proteins that are known to (a) bind to either DNA or RNA and (b) regulate core cellular processes like DNA repair, transcription, and translation.
Validating antibody specificity for immunohistochemistry

collaboration with Dana García (TxSt Department of Biology)
Historically, the zebrafish research community did not have a large set of antibodies raised against zebrafish proteins to use in immunoassays like Western blots and immunohistochemistry. In recent years, this scenario has changed as companies have developed and validated antibodies specifically for zebrafish proteins. However, a large body of work using zebrafish has been performed using antibodies raised against non-zebrafish proteins (such as human or mouse). We are working with Dr. García to validate those antibodies and identify which gene product(s) react with those antibodies, to better interpret Western blots and IHC images of zebrafish tissue or extracts. A paper describing our methods for screening putative epitopes has been published in Zebrafish.
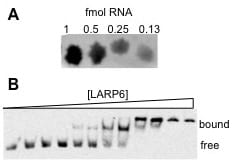
Agarose gel electrophoretic mobility shift assays
collaboration with L. Kevin Lewis (Department of Chemistry and Biochemistry)
We have combined recent improvements in native polyacrylamide gel shifts (Altschuler et al 2013) and agarose gel electrophoresis (Sanderson et al 2014) to develop a native agarose gel shift protocol using only standard molecular biology reagents. When a model protein:RNA system was quantitatively evaluated using this novel method, we obtained KD,app values that were comparable to a rapid polyacrylamide system. This study was published in Analytical Biochemistry.